Nifti objects data type¶
Another option for plotting and importing/exporting data is using the
Nifti objects. Nifti objects are a subclass of the neuroimaging format
Nibabel Nifti1Image, which is a file that generally has the
extension “.nii” or “.nii.gz”. This allows the user to use our methods
with the Nifti class but also use other functionality such as
Nibabel and Nilearn methods.
Load in the required libraries¶
import warnings
warnings.simplefilter("ignore")
%matplotlib inline
import supereeg as se
First, let’s load in an example nifti file, example_nifti:
nii = se.load('example_nifti')
Initialize nifti objects¶
Nifti objects can be initialized by passing any of the following to
the Nifti class instance initialization function: - A path to a
nifti file (ending in .nii or .nii.gz) - An existing Nifti object (this
makes a copy of the object) - A path to any other toolbox-supported
datatype, or an instance of another supported datatype (Brain or
Model objects)
You may also initialize a Nifti object using the load function
by specifying return_type='nii'.
For example:
bo_nii = se.Nifti('example_data')
Or:
bo_nii = se.load('example_data', return_type='nii')
Spatial resampling¶
Any Nifti object may be quickly resampled to an arbitrary voxel size
using the vox_size argument. The voxel sizes may be specified either
as a scalar (for cubic voxels) or as a 3D tuple (for rectangular prism
or parallelopiped voxels):
bo_nii = se.Nifti('example_data', vox_size=6)
Nifti object methods¶
Some useful methods on a nifti object:
nifti.info()¶
This method will give you a summary of the nifti object:
nii.info()
Header: <class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type :
db_name :
extents : 0
session_error : 0
regular :
dim_info : 0
dim : [ 4 30 36 30 500 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : float64
bitpix : 64
slice_start : 0
pixdim : [1. 6. 6. 6. 1. 1. 1. 1.]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 0
slice_code : unknown
xyzt_units : 0
cal_max : 0.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 0
glmin : 0
descrip :
aux_file :
qform_code : unknown
sform_code : aligned
quatern_b : 0.0
quatern_c : 0.0
quatern_d : 0.0
qoffset_x : -88.0
qoffset_y : -124.0
qoffset_z : -70.0
srow_x : [ 6. 0. 0. -88.]
srow_y : [ 0. 6. 0. -124.]
srow_z : [ 0. 0. 6. -70.]
intent_name :
magic : n+1
nifti.get_slice()¶
This method allows you to slice out images from your nifti object, and returns the indexed nifti.
nii_sliced = bo_nii.get_slice(index=[0,1,2])
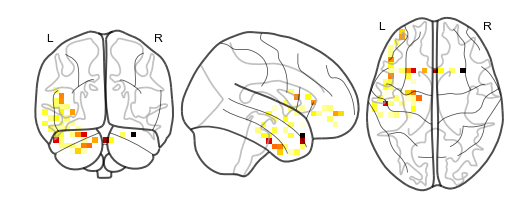
nifti.plot_glass_brain()¶
This method will plot your nifti object.
This method wraps nilearn.plot_glass_brain to plot the nifti object,
so any arguments that nilearn.plot_glass_brain accepts are supported
by nifti.plot_glass_brain.
nii_sliced.plot_glass_brain()
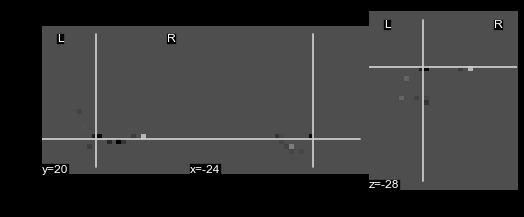
nifti.plot_anat()¶
This method will plot your nifti object.
This method wraps nilearn.plot_anat to plot the nifti object, so any
arguments that nilearn.plot_anat accepts are supported by
nifti.anat. For example, you can plot the example nifti:
nii.plot_anat()
nifti.make_gif()¶
This method will plot 4D nifti data as nilearn.plot_glass_brain,
save as png files, and compile the files as gif.
This method wraps nilearn.plot_glass_brain to plot the nifti object,
so any arguments that nilearn.plot_glass_brain accepts are supported
by nifti.plot_glass_brain.
#nii.make_gif(gifpath='/path/to/save/gif', index=range(0, 10), name=None, **kwargs)
nifti.save()¶
This method will save your nifti object to the specified filepath location as a ‘nii’ file.
#nii.save(filepath='/path/to/save/nifti')