Simulating a brain object¶
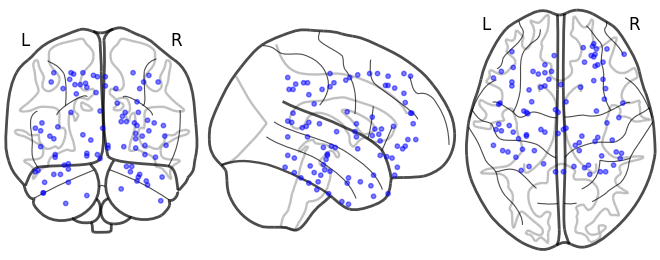
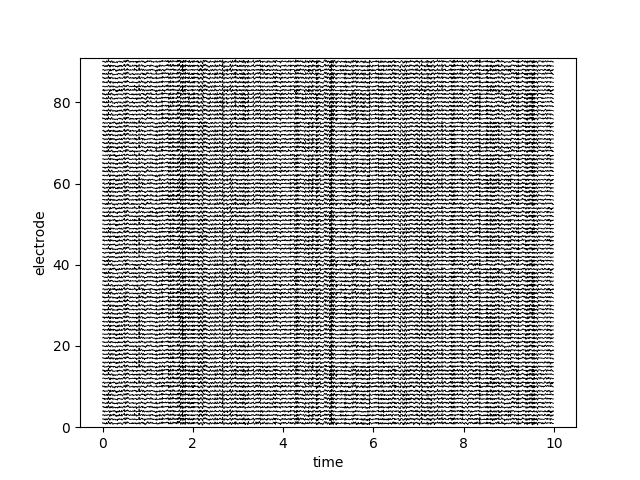
In this example, we demonstrate the simulate_bo function. First, we’ll load in some example locations. Then we’ll simulate 1 brain object specifying a noise parameter and the correlational structure of the data (a toeplitz matrix). We’ll then subsample 10 locations from the original brain object.
# Code source: Lucy Owen & Andrew Heusser
# License: MIT
import supereeg as se
from supereeg.helpers import _corr_column
import numpy as np
# simulate 100 locations
locs = se.simulate_locations(n_elecs=100)
# simulate brain object
bo = se.simulate_bo(n_samples=1000, sample_rate=100, cov='random', locs=locs, noise =.1)
# sample 10 locations, and get indices
sub_locs = locs.sample(90, replace=False).sort_values(['x', 'y', 'z']).index.values.tolist()
# index brain object to get sample patient
bo_sample = bo[: ,sub_locs]
# plot sample patient locations
bo_sample.plot_locs()
# plot sample patient data
bo_sample.plot_data()
# make model from brain object
r_model = se.Model(data=bo, locs=locs)
# predict
bo_s = r_model.predict(bo_sample, nearest_neighbor=False)
# find indices for reconstructed locations
recon_labels = np.where(np.array(bo_s.label) != 'observed')
# find correlations between predicted and actual data
corrs = _corr_column(bo.get_data().as_matrix(), bo_s.get_data().as_matrix())
# index reconstructed correlations
corrs[recon_labels].mean()
Total running time of the script: ( 0 minutes 0.834 seconds)