Predict unknown location¶
In this example, we load in a single subject example, remove electrodes that exceed a kurtosis threshold, load a model, and predict activity at all model locations and plot those locations. We then convert the reconstruction to a nifti and plot the reconstruction.
Out:
Number of electrodes: 274
Recording time in seconds: [ 5.3984375 14.1328125]
Sample Rate in Hz: [256, 256]
Number of sessions: 2
Date created: Wed Jul 25 20:26:51 2018
Meta data: {}
# Code source: Lucy Owen & Andrew Heusser
# License: MIT
import supereeg as se
# load example data
bo = se.load('example_data')
# load example model
model = se.load('example_model')
# the default will replace the electrode location with the nearest voxel and reconstruct at all other locations
reconstructed_bo = model.predict(bo, force_update=True)
# plot locations colored by label
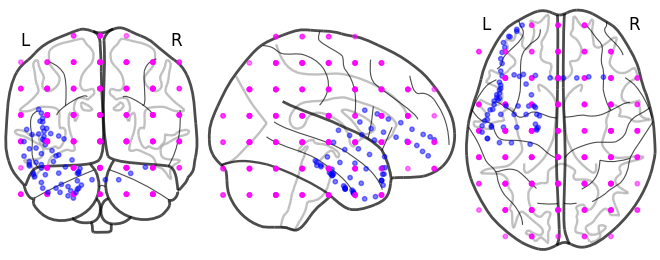
reconstructed_bo.plot_locs()
# print out info on new brain object
reconstructed_bo.info()
# save as nifti
reconstructed_nii = reconstructed_bo.to_nii(template='gray', vox_size=20)
# plot nifti reconstruction
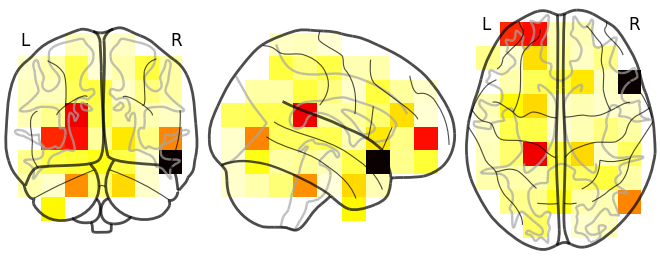
reconstructed_nii.plot_glass_brain()
Total running time of the script: ( 0 minutes 32.736 seconds)